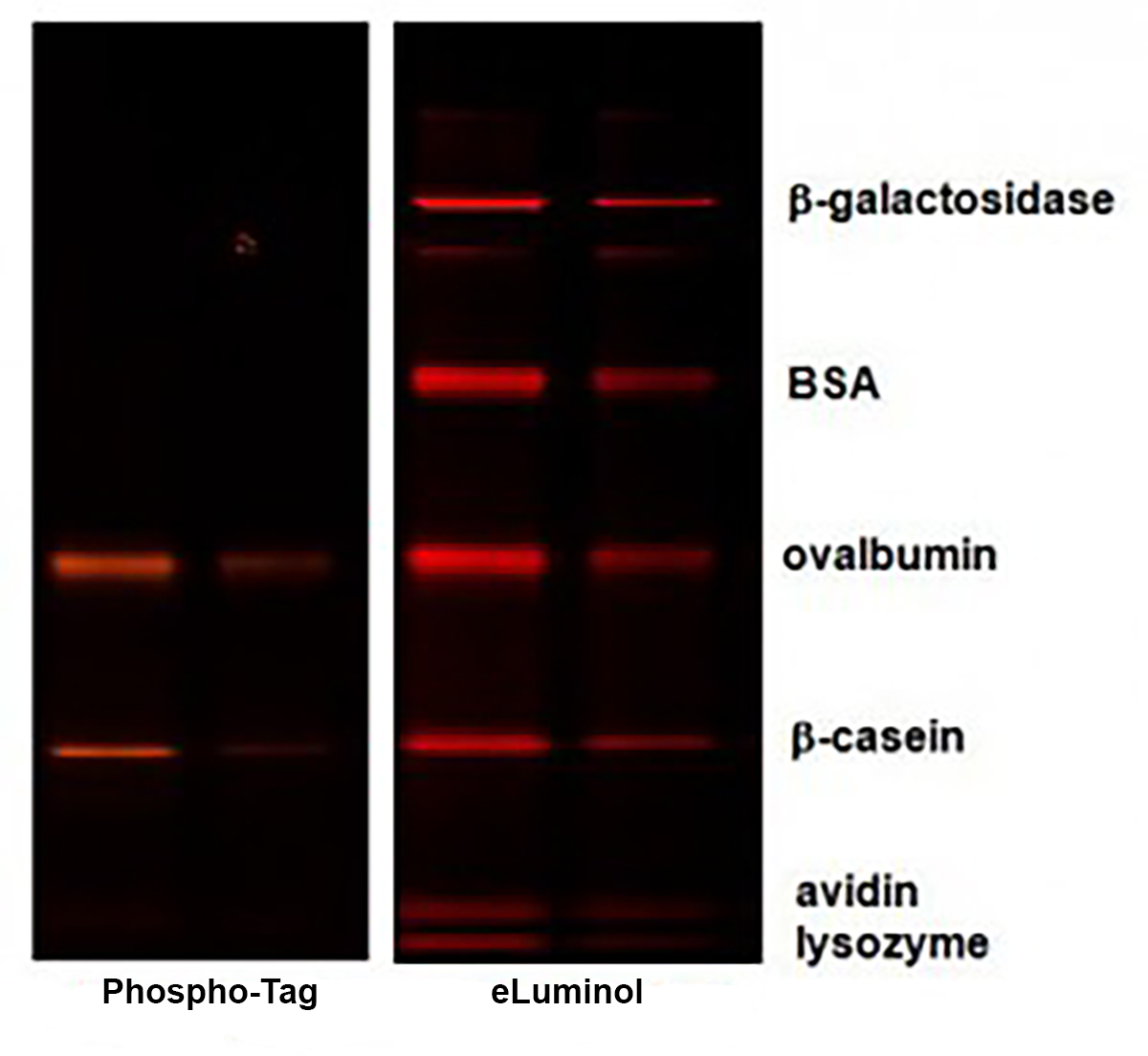
Phospho-Tag™ Phosphoprotein Gel Stain is a high sensitive fluorescent stain designed for selectively detecting phosphoproteins in polyacrylamide gels. This stain contains a phospho-tag™ group, which allows direct, in-gel detection of phosphate groups attached to tyrosine, serine, or threonine residues, without the need for antibodies or radioisotopes. The stain can be used with standard SDS-polyacrylamide gels or with 2-D gels.
Protein gel stain
Protein Targets of Inositol Pyrophosphate (5-IP7) in the parasite Trypanosoma cruzi
cMyBPC phosphorylation modulates the effect of omecamtiv mecarbil on myocardial force generation
In-Depth Characterization of the Clostridioides difficile Phosphoproteome to Identify Ser/Thr Kinase Substrates
Trypanosoma cruzi DNA Polymerase ? Is Phosphorylated In Vivo and In Vitro by Protein Kinase C (PKC) and Casein Kinase 2 (CK2)
Ser/Thr kinase-dependent phosphorylation of the peptidoglycan hydrolase CwlA controls its export and modulates cell division in Clostridioides difficile
Affinity-based proteomics reveals novel targets of inositol pyrophosphate (5-IP7)-dependent phosphorylation and binding in Trypanosoma cruzi replicative stages
Identification of Phosphorylation and Other Post-Translational Modifications in the Central C4C5 Domains of Murine Cardiac Myosin Binding Protein C
Increased phosphorylation of HexM improves lysosomal uptake and potential for managing GM2 gangliosidoses
Reversible phosphorylation of cyclin T1 promotes assembly and stability of P-TEFb
Substrate Profiling of the Arabidopsis Ca2+-Dependent Protein Kinase Atcpk4 and its Ricinus Communis Ortholog Rccdpk1
Serine phosphorylation regulates the P-type potassium pump KdpFABC
Applications
Protocol (PDF):
P005A
MSDS (PDF):
P005A
Cited Reference:
Brian S. Mantilla, Nathaniel Brown, Dorothea Fiedler, Roberto Docampo
bioRxiv 2020.01.09.900860; doi: https://doi.org/10.1101/2020.01.09.900860
Ranganath Mamidi, Joshua B. Holmes, Chang Yoon Doh, Katherine L. Dominic, Nikhil Madugula, and Julian E. Stelzer
J. Gen. Physiol. 2021, 153, 7, e202012816. https://doi.org/10.1085/jgp.202012816
Transito Garcia-Garcia, Thibaut Douche, Quentin Giai Gianetto, Sandrine Poncet, Nesrine El Omrani, Wiep Klaas Smits, Elodie Cuenot, Mariette Matondo, and Isabelle Martin-Verstraete
Mol Cell Proteomics, 2022, 21(11), 100428. DOI:https://doi.org/10.1016/j.mcpro.2022.100428
Maldonado, E.; Rojas, D.A.; Urbina, F.; Valenzuela-Pérez, L.; Castillo, C.; Solari, A.
Cells 2022, 11, 3693. https://doi.org/10.3390/cells11223693
Garcia-Garcia T, Poncet S, Cuenot E, Douché T, Giai Gianetto Q, Peltier J, Courtin P, Chapot-Chartier M-P, Matondo M, Dupuy B, Candela T, Martin-Verstraete I.
mBio 12:e00519-21. https://doi.org/10.1128/mBio.00519-21.
Mantilla, BS, Kalesh, K, Brown, NW, Fiedler, D, Docampo, R.
Mol Microbiol. 2021; 115: 986–1004. https://doi.org/10.1111/mmi.14672
Chang Yoon Doh, Katherine L. Dominic, Caitlin E. Swanberg, Nikhil Bharambe, Belinda B. Willard, Ling Li, Rajesh Ramachandran, and Julian E. Stelzer
ACS Omega, 2022, 7, 16, 14189–14202. https://doi.org/10.1021/acsomega.2c00799
Graeme Benzie, Kristen Bouma, Taylor Battellino, Steven Cooper, Rick Hemming, Wafa Kammouni, Lin Liu, Cuong Do, Mazdak Khajehpour, Helene Perreault, Stuart Kornfeld, Barbara Triggs-Raine, Brian L. Mark
BBA Advances, 2022, 2, 100032, https://doi.org/10.1016/j.bbadva.2021.100032.
Fang Huang, Trang TT Nguyen, Ignacia Echeverria, Ramachandran Rakesh, Daniele C Cary, Hana Paculova, Andrej Sali, Arthur Weiss, Boris Matija Peterlin, Koh Fujinaga
eLife 10:e68473. https://doi.org/10.7554/eLife.68473
Kilburn, Ryan and Fedosejevs, Eric T. and Mehta, Devang and Ghahremani, Mina and Thelen, Jay J. and Uhrig, Richard Glen and Snedden, Wayne A. and Plaxton, William Charles
SSRN: http://dx.doi.org/10.2139/ssrn.4238554
Marie E SweetXihui ZhangHediye Erdjument-BromageVikas DubeyHimanshu KhandeliaThomas A NeubertBjørn P PedersenDavid L Stokes
eLife, 2020, 9:e55480. https://doi.org/10.7554/eLife.55480
Phospho-Tag™ Phosphoprotein Gel Stain
Introduction
To Order
Documents
Contact Us
ABP Biosciences
405 E Gude Dr, STE 214
Rockville, MD 20850
Service Hotline
Tel: 301-658-7993
E-mail: info@abpbio.com
Copyright@ 2026 ABP Biosciences, LLC. All Rights Reserved.